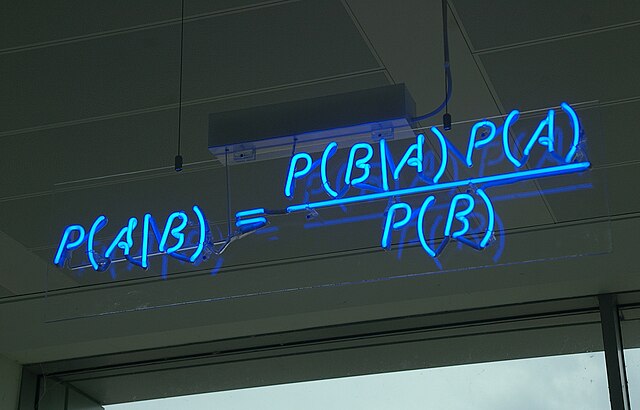 If I said you had a nice posterior Reverend Bayes, would you take offense? Photo via WikiMedia Commons.
If I said you had a nice posterior Reverend Bayes, would you take offense? Photo via WikiMedia Commons.Over at The Molecular Ecologist, new contributor Peter Fields—a Ph.D. student studying plant-pathogen coevolution at the University of Virginia—writes about approximate Bayesian computation and a new approach to this still-developing method of statistical inference that can make it quite a bit faster.
ABC functions upon the rationale that the likelihood might be approximated through the use of simulation and simulation summary statistics2, and that the evaluation of model fit to a dataset can be identified through a comparison of Ss derived from simulated scenarios and calculation of those same summaries on an observed, empirical dataset. In theory, simulation summaries are selected to provide maximal distinction amongst competing models. In practice, identifying these summaries isn’t always easy, and is the object of continued research3
For an introduction to ABC, and a description of the new approach, go read the whole thing.◼



